Erwin Chargaff was the first to notice, in the early 1950s, before Watson and Crick deduced the structure of DNA, that the quantity of purines in DNA equals the quantity of pyrimidines (specifically, the amount of adenine equals the amount of thymine; and the amount of guanine equals the amount of cytosine). This observation was key to establishing the structure of DNA, and it is often cited as Chargaff's first parity rule. But Chargaff also made another observation (the second parity rule), namely that even within a
single strand of DNA, the amount of adenine tends to equal the amount of thymine and the amount of guanine tends to equal the amount of cytosine.
It's easy to understand why the first parity rule holds true, because complementarity of DNA strands depends on A pairing with T and G pairing with C; these pairings give rise to the "rungs" of the DNA ladder and ensure that copying of strands occurs with total fidelity during cell division. But there doesn't seem to be any
a priori reason why the second parity rule should hold true. And in fact, it often
doesn't hold true, as
Wacław Szybalski noted in 1966 when he
reported finding imbalances of purines and pyrimidines in bacteriophage and other DNA samples. Szybalski observed that in most cases, protein-coding regions of DNA tend to have slightly more purines than pyrimidines on one strand and slightly more pyrimidines than purines on the other strand, such that messenger RNA ends up purine-heavy.
If you're having trouble visualizing the situation, imagine a very short (12-base) "chromosome" containing 50% G+C content. One possibility is that one strand looks like GGGGGGTTTTTT and the other strand is CCCCCCAAAAAA. In this case half the purines (all the G's) are on one strand and half (A's) are on the other. But you could just as easily have strands be GGGGGGAAAAAA and CCCCCCTTTTTT. In this case, one strand is all-purines, the other all-pyrimidines. Both examples violate Chargaff's second rule, which requires that G = C and A = T within each strand (e.g., GGGCCCTTTAAA + CCCGGGAAATTT would obey the rule).
To my knowledge, no one has yet reported the fact (which I'll now report) that the degree to which Chargaff's second parity rule is violated depends on the G+C content of the source genome (at least for bacteria). Simply put, organisms with a G+C content of around 68% obey Chargaff's rules. Organisms with more than 68% G+C content violate Chargaff's second rule in the direction of
pyrimidine loading of mRNA. Organisms with
less than 68% G+C content (which of course includes the overwhelming majority of organisms) have purine-heavy DNA, to a degree that depends on the amount of A+T in the DNA.
 |
| Purine/pyrimidine ratio (in coding regions) as a function of genome G+C content based on codon analysis of 93 organisms. As genomes become more A+T rich, mRNA becomes more heavily purine-loaded. |
The above graph shows how this relationship works. To create the graph, I did a statistical analysis of codon usage in 93 bacterial species. Organisms were chosen so as to obtain representatives across the AT/GC spectrum. No genus is represented more than once. In order to get as broad a sampling as possible, I included 14 intracellular symbionts with ultra-low G+C content (plus one such creature—
Candidatus Hodgkinia cicadicola—with a 58% G+C content); many extremophiles; heterotrophs and autotrophs; pathogens and non-pathogens; and organisms with large and small genomes. The complete organism list is presented in a table further below.
Codon usage statistics for each organism were obtained using tools at
http://genomevolution.org. Relative prevalences of A, T, G, and C in the genomes' coding regions were determined by codon frequency analysis. The purine:pyrimidine ratio was simply calculated as (A+G)/(C+T) based on the codon-wise frequency of usage of each base.
What we see is that while there is a good deal of noise in the data, nevertheless it's quite clear that purine/pyrimidine ratios increase sharply as genome G+C decreases.Organisms for which Chargaff's second rule holds true (points falling at y = 1.0) are in a small minority. Most organisms have purine-rich coding regions, resulting in purine-rich mRNA.
Purine enrichment occurs for both adenine and guanine. For example, in
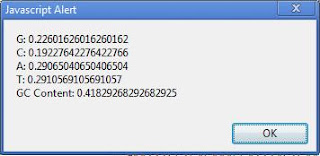
Clostridium botulinum (genome G+C = 28.21%), codon analysis reveals G/C/A/T relative abundances (on the coding strand) of 18.3/10.8/40.3/30.6.
Intra-codon base position analysis reveals that purine enrichment is far more concentrated in position one of the codon than other positions. The graphs below show the purine balance on a position-by-position basis, for each base in a codon.
Most of the variation in purine/pyrimidine ratio happens in position 1 of the codon (the 'A' in ATG, for example). Notice that the purine/pyrimidine ratio in this position is well above 1.0 for all organisms.
Variation in purine loading at the second position of the codon is more carefully controlled (notice that there is less "scatter" in this graph). The y-axis scale is different here than in the previous graph, hence the slope is quite a bit less pronounced than it looks. Also, notice that most of the points in this plot are below parity (i.e., below 1.0 on the y-axis), indicating that this codon position is relatively pyrimidine-rich.
The third (so-called "wobble") position of the codon shows considerable variation in values, but the slope of the curve is less than in the previous two graphs, and this position is pyrimidine-rich for about two-thirds of the organisms.
It's well known that GC-skew tends to be exaggerated in position 3 of the codon. For example, if the overall genome G+C is 70%, the position-wise G+C for the wobble base may be 90%. Surprisingly, we find that purine loading is most exaggerated in position 1 of the codon, not position 3. Not only is the slope of the purine-ratio curve shallower in position 3 than for the other two base positions, only position 1 is actually purine-heavy: positions 2 and 3 tend to be net pyrimidine-rich. This fact (that purine loading is primarily localized to codon position 1, whereas GC-skew is exaggerated in position 3) might indicate that the forces responsible for purine loading are entirely different from the forces responsible for GC skew.
What might those forces be? What kinds of selection pressure might cause organisms to purine-load one strand of their DNA? One possibility is that purine loading of the coding strand is a strategy for protecting the "weaker" or more vulnerable strand from damage or mutations. Cytosine is thought to be particularly vulnerable to deamination (and later substitution with thymine, during repair). It's possible that the transcription process (which is
asymmetric, in that RNA polymerase operates against just one strand of DNA, leaving the other strand free) is protective of the antisense strand of DNA. That is, in transcription, RNA polymerase cloaks the antisense strand and in so doing renders that strand less vulnerable to deamination events, rogue methylations, etc., while transcription is taking place.
An entirely different possibility is envisioned by an RNA World hypothesis. In this hypothesis, the genetic material of early ancestor organisms was single-stranded RNA. Since single-stranded RNA is not "complementary" to anything, there is no need for it to obey Chargaff symmetries. Thus, purine loading could have occurred prior to the advent of double-stranded DNA, and early organisms could have been uniformly AT-rich. In this model of the world, GC-rich genomes are a late development, and the processes responsible for creating GC-rich DNA led to genetic material with full Chargaff base parity.
We may not know for a long time (if ever) what the mechanisms of purine enrichment are. But we know for sure that purine accumulation is a widespread phenomenon in the bacterial world (operating across diverse clades) and happens in a way that encourages purine-rich mRNA in organisms with low G+C content in their genomes.
Organisms used in this study:
| Organism |
GC% |
genome size |
| Anaeromyxobacter dehalogenans
2CP-1 |
74.67 |
5009007 |
| Cellulomonas flavigena strain
DSM 20109 |
74.29 |
4123179 |
| Xylanimonas cellulosilytica
strain DSM 15894 |
72.47 |
3831380 |
| Streptomyces
bingchenggensis strain BCW-1 |
70.75 |
11936683 |
| Myxococcus fulvus strain HW-1 |
70.63 |
9003593 |
| Rubrobacter
xylanophilus strain DSM 9941 |
70.48 |
3225748 |
| Rhodospirillum centenum ATCC
51521 |
70.46 |
4355543 |
| Actinomyces sp. oral taxon 175
strain F0384 |
68.73 |
3133330 |
| Rhodococcus
equi strain ATCC 33707 |
68.72 |
5259057 |
| Acidovorax
avenae subsp. citrulli strain AAC00-1 |
68.53 |
5352772 |
| Bordetella bronchiseptica
strain RB50 |
68.08 |
5339179 |
| Alicycliphilus denitrificans
strain K601 |
67.81 |
5070751 |
| Stenotrophomonas
maltophilia strain JV3 |
66.89 |
4544477 |
| Rhodobacter
capsulatus strain SB 1003 |
66.56 |
3871920 |
| Pseudomonas
aeruginosa strain PA7 |
66.45 |
6588339 |
| Ralstonia
eutropha strain H16 |
66.29 |
7416678 |
| Xanthomonas campestris pv.
raphani strain 756C |
65.29 |
4941214 |
| Thioalkalivibrio
sp. strain HL-EbGR7 |
65.06 |
3470516 |
| Rhodopseudomonas
palustris strain BisB18 |
64.96 |
5513844 |
| Brevundimonas
diminuta strain ATCC 11568 |
64.51 |
3369316 |
| Rhodothermus
marinus strain DSM 4252 |
64.09 |
3386737 |
| Bradyrhizobium
japonicum strain USDA 110 |
64.06 |
9105828 |
| Mycobacterium
tuberculosis strain C |
63.82 |
4379118 |
| Thermanaerovibrio
acidaminovorans strain DSM 6589 |
63.79 |
1848474 |
| Halomonas elongata DSM 2581
strain type DSM 2581 |
63.61 |
4061296 |
| Novosphingobium nitrogenifigens
strain DSM 19370 |
63.43 |
4182647 |
| Polaromonas sp. strain JS666
|
62.24 |
5898676 |
| Desulfovibrio africanus strain
Walvis Bay |
61.42 |
4200534 |
| Candidatus Desulforudis
audaxviator strain MP104C |
60.85 |
2349476 |
| Burkholderia rhizoxinica strain
HKI 454 |
60.68 |
3750138 |
| Slackia heliotrinireducens
strain DSM 20476 |
60.21 |
3165038 |
| Candidatus Nitrospira defluvii |
59.03 |
4317083 |
| Halogeometricum borinquense DSM
11551 |
58.43 |
3944467 |
| Candidatus Hodgkinia cicadicola
strain Dsem |
58.39 |
143795 |
| Sideroxydans lithotrophicus
strain ES-1 |
57.54 |
3003656 |
| Cenarchaeum symbiosum A |
57.37 |
2045086 |
| Serratia sp. strain AS12 |
55.96 |
5443009 |
| Acidaminococcus
fermentans strain DSM 20731 |
55.84 |
2329769 |
| Hyperthermus butylicus strain
DSM 5456 |
53.74 |
1667163 |
| Methanosaeta thermophila
(Methanothrix thermophila PT) strain PT |
53.55 |
1879471 |
| Neisseria gonorrhoeae strain
NCCP11945 |
53.37 |
2236178 |
| Treponema paraluiscuniculi
strain Cuniculi A |
52.74 |
1133390 |
| Pseudovibrio sp. strain FO-BEG1 |
52.38 |
5916782 |
| Nitrosococcus halophilus strain
Nc4 |
51.60 |
4145260 |
| Herpetosiphon
aurantiacus DSM 785 |
50.84 |
6785430 |
| Escherichia coli B strain
REL606 |
50.77 |
4629812 |
- Bdellovibrio
bacteriovorus strain ATCC15356;
|
50.65 |
3782950 |
| Pectobacterium
wasabiae strain WPP163 |
50.48 |
5063892 |
| Anaplasma
centrale (Anaplasma marginale subsp. centrale str. Israel) strain
Israel |
49.98 |
1206806 |
| Actinomyces
coleocanis strain DSM 15436 |
49.47 |
1723843 |
| Desulfotalea
psychrophila strain LSv54 |
46.72 |
3659634 |
| Polynucleobacter
necessarius strain STIR1 |
45.56 |
1560469 |
| Nitrosomonas sp. strain Is79A3 |
45.44 |
3783444 |
| Coprothermobacter
proteolyticus strain DSM 5265 |
44.77 |
1424912 |
| Vibrio
sp. Ex25 strain EX25 |
44.57 |
5160431 |
| Geobacillus
thermoglucosidans strain TNO-09.020 |
43.82 |
3740238 |
| Waddlia chondrophila strain
2032/99 |
43.59 |
2139757 |
| Bacteroides fragilis strain
638R |
43.42 |
5373121 |
| Thiomicrospira
crunogena strain XCL-2 |
43.13 |
2427734 |
| Coxiella
burnetii strain CbuG_Q212 |
42.63 |
2008870 |
| Chlamydia muridarum Nigg strain
MoPn |
40.27 |
1080451 |
| Psychromonas ingrahamii strain
37 |
40.09 |
4559598 |
| Nitratiruptor sp. strain
SB155-2 |
39.69 |
1877931 |
| Lactobacillus reuteri strain
DSM 20016 |
38.87 |
1999618 |
| Thermotoga lettingae strain TM |
38.70 |
2135342 |
| Streptococcus
pyogenes strain Alab49 |
38.63 |
1841271 |
| Bartonella bacilliformis strain
ATCC 35685; KC583 |
38.24 |
1445021 |
| Halothermothrix
orenii strain DSM 9562; H 168 |
37.78 |
2463968 |
| Staphylothermus marinus strain
F1 |
35.73 |
1570485 |
| Calditerrivibrio nitroreducens
strain DSM 19672 |
35.69 |
2216552 |
| Bacillus
thuringiensis serovar andalousiensis strain BGSC 4AW1 |
34.96 |
5488844 |
| Desulfurobacterium
thermolithotrophum |
34.95 |
1541968 |
| Wolbachia
pipientis strain wPip |
34.19 |
1482455 |
| Nitrosopumilus maritimus strain
SCM1 |
34.17 |
1645259 |
| Staphylococcus aureus strain
04-02981 |
32.90 |
2821452 |
| Methanobrevibacter ruminantium
strain M1 |
32.64 |
2937203 |
| Rickettsia
japonica strain YH |
32.35 |
1283087 |
| Methanocaldococcus fervens
strain AG86 (v1)
|
32.21 |
1507251 |
| Mycoplasma
genitalium G37 strain G-37 |
31.69 |
580076 |
| Nanoarchaeum equitans strain
Kin4-M |
31.56 |
490885 |
| Orientia tsutsugamushi strain
Boryong |
30.53 |
2127051 |
| Methanococcus aeolicus strain
Nankai-3 |
30.04 |
1569500 |
| Candidatus
Pelagibacter ubique strain HTCC1062 |
29.68 |
1308759 |
| Ehrlichia
canis strain Jake |
28.96 |
1315030 |
| Arcobacter nitrofigilis strain
DSM 7299 |
28.36 |
3192235 |
| Clostridium
botulinum A strain ATCC 19397 |
28.21 |
3863450 |
| Parvimonas
sp. oral taxon 393 strain F0440 |
28.17 |
1483165 |
| Candidatus Arthromitus sp.
strain SFB-mouse-NYU |
27.94 |
1569870 |
| Candidatus Blochmannia
floridanus |
27.38 |
705557 |
| Buchnera aphidicola
(Acyrthosiphon pisum) strain 5A |
25.69 |
653223 |
| Wigglesworthia glossinidia
endosymbiont of Glossina brevipalpis |
22.48 |
703004 |
| Candidatus
Sulcia muelleri strain CARI (v1)
|
21.13 |
276511 |
| Candidatus Carsonella ruddii
strain PV (v1) |
16.56 |
159662 |